Benchmarking LLMs for global health
A dataset and benchmarking pipeline using synthetic personas to understand and optimize LLM performance for tropical and infectious diseases (TRINDs).
Quick links
Large language models (LLMs) have shown potential for medical and health question-answering across various health-related tests and spanning different formats and sources. Indeed we have been on the forefront of efforts to expand the utility of LLMs for health and medical applications, as demonstrated in our recent work on Med-Gemini, MedPaLM, AMIE, Multimodal Medical AI, and our release of novel evaluation tools and methods to assess model performance across various contexts. Especially in low-resource settings, LLMs can potentially serve as valuable decision-support tools, enhancing clinical diagnostic accuracy, accessibility, and multilingual clinical decision support, and health training, especially at the community level. Yet despite their success on existing medical benchmarks, there is still some uncertainty about how well these models generalize to tasks involving distribution shifts in disease types, region-specific medical knowledge, and contextual variations across symptoms, language, location, linguistic diversity, and localized cultural contexts.
Tropical and infectious diseases (TRINDs) are an example of such an out-of-distribution disease subgroup. TRINDs are highly prevalent in the poorest regions of the world, affecting 1.7 billion people globally with disproportionate impacts on women and children. Challenges in preventing and treating these diseases include limitations in surveillance, early detection, accurate initial diagnosis, management, and vaccines. LLMs for health-related question answering could potentially enable early screening and surveillance based on a person’s symptoms, location, and risk factors. However, only limited studies have been conducted to understand LLM performance on TRINDs with few datasets existing for rigorous LLM evaluation.
To address this gap, we have developed synthetic personas — i.e., datasets that represent profiles, scenarios, etc., that can be used to evaluate and optimize models — and benchmark methodologies for out-of-distribution disease subgroups. We have created a TRINDs dataset that consists of 11,000+ manually and LLM-generated personas representing a broad array of tropical and infectious diseases across demographic, contextual, location, language, clinical, and consumer augmentations. Part of this work was recently presented at the NeurIPS 2024 workshops on Generative AI for Health and Advances in Medical Foundation Models.
TRINDs dataset development and benchmarking overview.
Creating synthetic TRINDs personas to evaluate LLMs
We examined authoritative sources, including the WHO, PAHO and CDC, that publish factual information about different diseases, and used what we learned to create an initial seed set of patient persona templates for each disease. These personas consist of general symptoms, direct attributes, and specific symptoms. They also include context, lifestyle, and risk factors that were reviewed by clinicians to confirm the accuracy and the clinical relevance of the formatting for the personas. These original seed personas currently span 50 diseases.
Building blocks for the TRINDs dataset.
We utilize LLM prompting to expand the seed set of synthetic personas to include demographic and semantic clinical and consumer augmentations (see below), resulting in a total of 11,000+ personas. Additionally, we manually translated the seed set into French to enable evaluation on how distribution shifts in language impact model performance. We then developed an LLM-based autorater that scores an answer as correct if the ground truth and predicted diagnosis are the same or meaningfully similar to each other.
Examples of an original seed persona and LLM-augmentations.
Evaluation
LLM performance on TRINDs vs USMLE
We evaluate the accuracy of Gemini models (Gemini 1.5) in identifying a disease indicated from persona descriptions. We demonstrate that there are distribution shifts in model performance on this dataset compared to USMLE-based benchmark datasets, with lower performance on the TRINDs set compared to reported performance on US datasets
The relevance of context
We systematically perform evaluations with the dataset to understand the impact of different contexts, types (clinical vs. consumer), demographics (age, race, gender), and semantic styles. We look at how combinations of symptoms, risk factors, location, and demographics impact LLM accuracy to provide an accurate diagnosis given full or partial context. Evaluations demonstrate that including location and risk factors in combination with specific and general symptoms results in the highest performance, suggesting that symptoms alone may be insufficient for accurate responses.
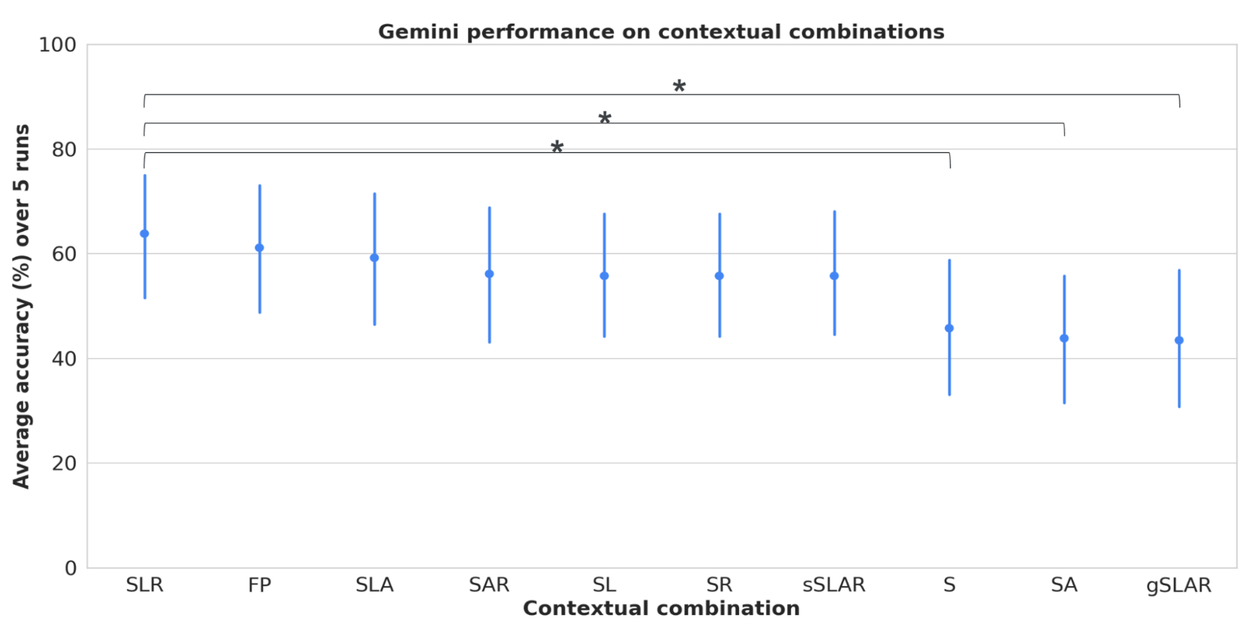
Model performance on contextual combinations of symptoms (general and specific), location, risk factors, and demographic attributes. S = symptoms, L = location, A = attributes, R = risk factors, gS = general symptoms, sS = specific symptoms, FP = full persona with all context. Error bars = 95% confidence interval. Note: FP accuracy is also significantly higher than S, SA and gSLAR.
Performance across race, gender, and counterfactual location
We examined the impact of including a statement identifying race, e.g.,“I am Black” or “Patient is racially Asian”. We also switched out gender references and evaluated the impact of using female, male and non-binary language on performance. We found no statistically significant difference in performance across race and gender. We also examine the impact of indicating low-incident locations (counterfactual location) on performance, described in the paper.
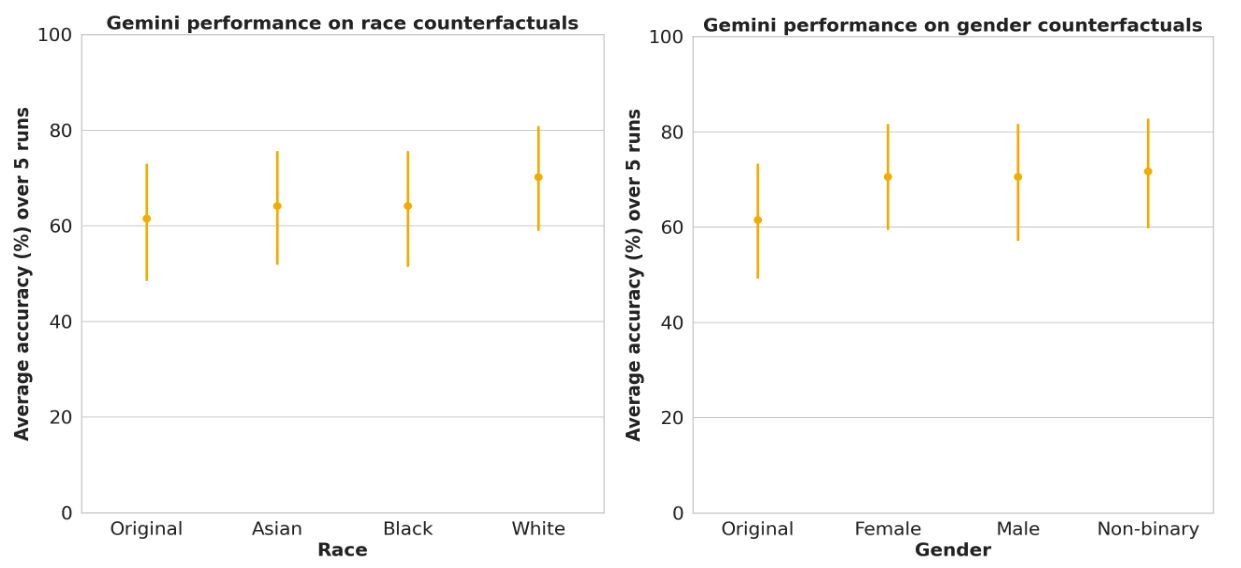
LLM performance on race and gender subgroups in TRINDs demonstrate no significant differences. Error bars = 95% confidence interval.
Human expert performance and evaluation
We recruited 7 experts with 10+ years of experience in TRINDs and public health to answer open-ended short answer questions (SAQ) and multiple choice questions (MCQ). We characterized the performance of the top five experts and also generated LLM performance on the same subset of data. This allowed simulation of different scenarios of expert round tables.
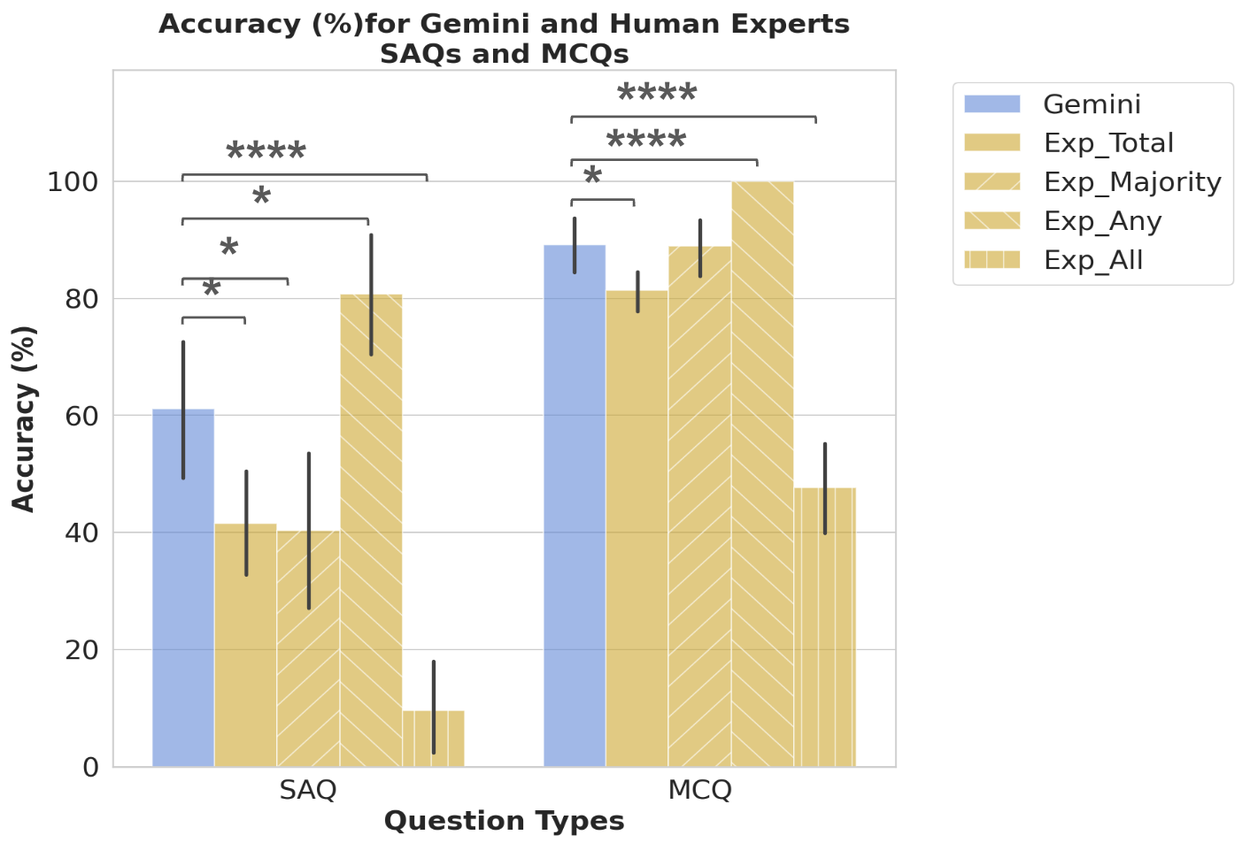
Comparison of LLM (Gemini) performance to that of the the top five experts, which were characterized with four scores: 1) the average across the total expert score (Exp_Total); 2) the full score if the majority vote was correct (Exp_Majority); 3) the full score if any expert was correct (Exp_Any); and 4) the full score only if all experts were correct (Exp_All), allowing us to explore a variety of expert decision making scenarios. Error bars = 95% confidence interval.
While LLMs performed worse on TRINDs than on USMLE, they performed better than the best performing expert, and most of the expert combination scenarios except the Exp_Any scenario, which is a more appropriate benchmark to beat as it reflects a round table of public health experts each with specific focal areas coming together to make decisions.
Per disease performance
We find that the LLMs tend to more accurately identify common diseases (e.g., HIV) or diseases with specific symptoms and risk factors (e.g., rabies). Certain diseases, such as tapeworm infection, are more susceptible to being labeled inaccurately if only symptoms are provided. Additionally, certain diseases, like rabies) are more robust to counterfactual locations than others.
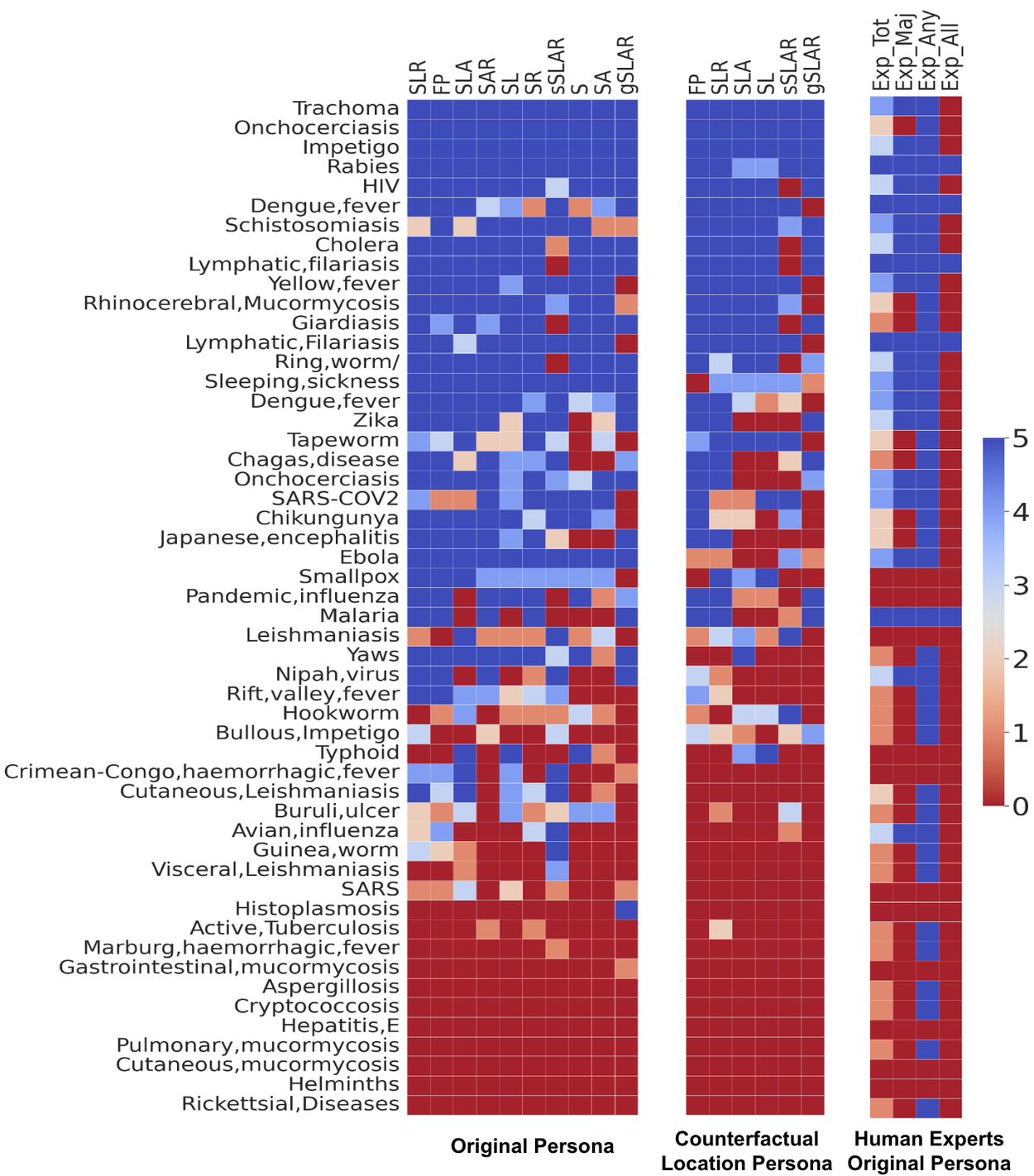
Per disease performance of the LLM on tropical and infectious diseases shown with different contextual combinations (left), contextual combinations with location counterfactual (center), and the human expert performance (right). Contextual combinations abbreviated as above.
Improving LLM performance through in-context learning
We tune the LLM (Gemini 1.5) through in-context learning with simple multi-shot prompting with the seed set of 50 questions (1 per disease) with all the symptoms, locations, and risk factors. This improves the performance on both demographic and semantic augmentations, demonstrating that this gap can be rectified with intentional tuning and that the seed datasets can be used to optimize LLM performance.
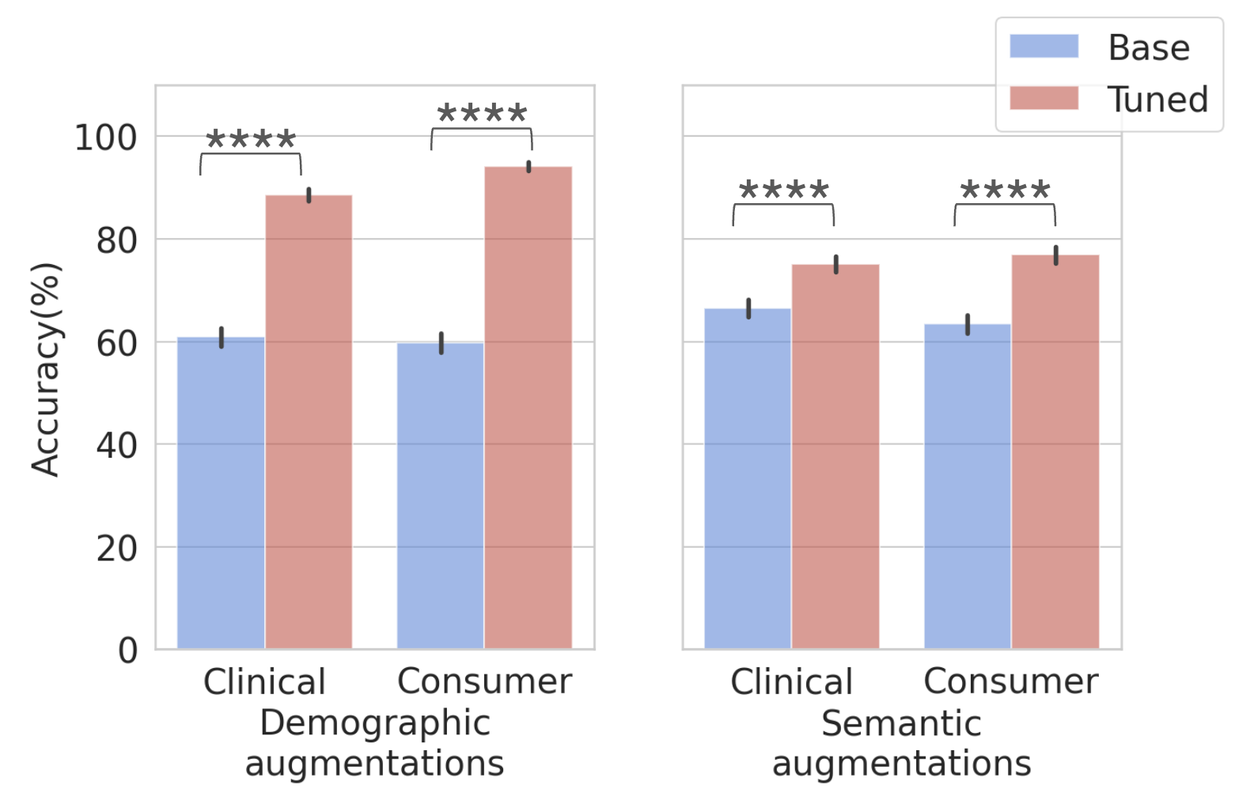
Performance on demographic (left) and semantic (right) augmented datasets before and after in-context tuning. Total = 10,570, clinical demographic (n=2635), consumer demographic (n=2635), clinical semantic (n=2650) and consumer semantic (n=2650). Error bars = 95% confidence interval.
Assessing potential for LLM-based tools for disease screening
We developed a TRINDs disease screening user interface, powered by a version of Gemini tuned for this task, that allows users to easily put in their demographics, location, lifestyle information, and risk factors, to then select their symptoms from a comprehensive list and be provided a diagnosis. We connected the tool to the Our World in Data API to display incidence rates.
User interface and use case of LLM-based tool for tropical and infectious disease screening.
At this early stage, expert ratings (shown below) suggest the interface, while seemingly simple in output, has the potential to be a highly impactful and user-friendly reference tool for infectious diseases, benefiting both clinicians and researchers.
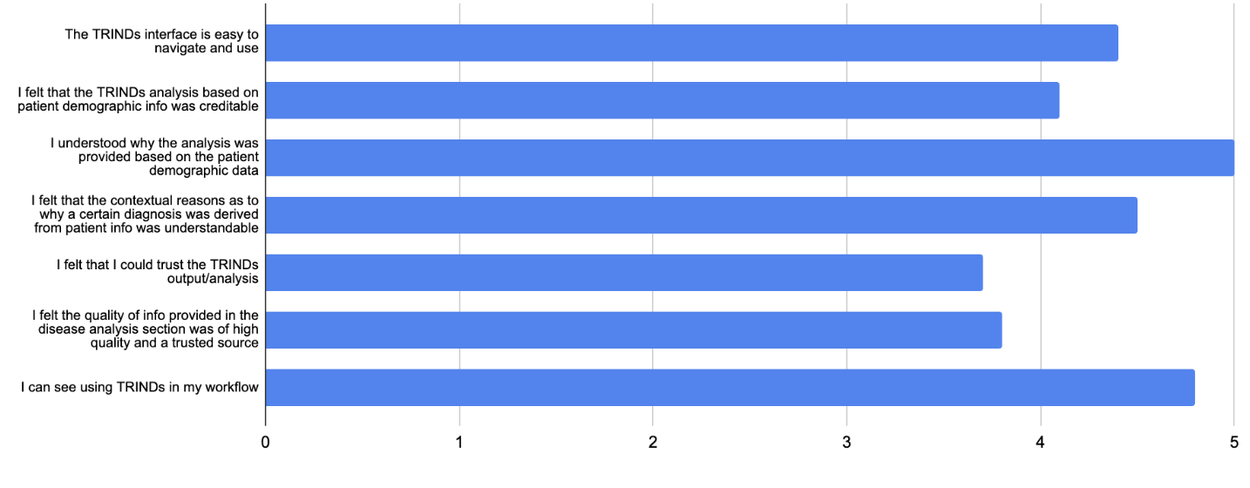
Expert ratings of TRINDs tool.
Implications
For healthcare workers, our findings highlight the potential of LLM-based tools to serve as valuable decision-support tools in resource-limited settings. However, these tools should complement, not replace, clinical judgment. They should be balanced by continuous assessment and updated regularly to accommodate temporal distribution shifts in real-world scenarios and reliability in diverse clinical settings. Given the sensitivity of health-related outcomes, it is essential that LLMs are evaluated for accurate, contextual, and culturally relevant performance. With these considerations in mind, translating the approach described here into a clinical tool would require further validation and standard regulatory review processes. Future research directions include expanding the benchmarks to encompass multilinguality and multimodality.
Acknowledgements
We would like to acknowledge the authors and contributors to this work: Mercy Asiedu, Nenad Tomasev, Tiya Tiyasirichokchai, Chintan Ghate, Awa Dieng, Katherine Heller, Mariana Perroni, Divleen Jeji, Heather Cole-Lewis. Thanks to our external experts Oluwatosin Akande, Geoffrey Siwo, Steve Adudans, Sylvanus Aitkins, Odianosen Ehiakhamen, and Eric Ndombi. Thanks to Marian Croak for her support and leadership.